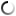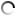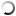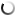Bacteria with chitinase activity were identified, and the diversity of chitinases contained in the genomes of related marine microorganisms was analyzed.
Students of the Department of Microbiology, Faculty of Biology, Moscow State University, during an internship at the White Sea Biological Station named after N.A. Pertsova became interested in bacteria that live in the White Sea and are capable of decomposing chitin. Chitin is a natural polymer similar to cellulose, but consisting of N-acetylglucosamine molecules, as a result of which its chains bind together, forming a strong structure with increased crystallinity. Chitin is very widespread on our planet, being in second place in quantity after cellulose. This is explained by the fact that it is a structure-forming element in the exoskeletons of invertebrate animals inhabiting the world’s oceans and fresh waters of the Earth, and is also present in the cell walls of fungi and the cuticle of arthropods. The decomposition of this substance is an important stage in the biogeochemical carbon cycle, causing the return of biogenic elements to it. It is caused by the action of microbial chitinases, and their composition varies depending on the place of isolation of chitinolytic microorganisms (terrestrial, freshwater, marine ecosystems). Chitin-degrading enzymes—chitinases—are in demand in various fields of biotechnology.
The purpose of the research was to study the diversity of chitinolytic bacteria living in the Kandalaksha Bay of the White Sea in the area of the White Sea Biological Station named after N.A. Pertsova. For this purpose, enrichment cultures of microorganisms using chitin as the only source of carbon and energy were obtained from samples of invertebrate animals, algae and water from the White Sea. The ability of enrichment cultures to decompose material was tested on Petri dishes with a medium containing chitin, where, as a result of the action of chitinases, zones of clearing of the initially turbid medium appeared. DNA was isolated from such cultures for amplification and subsequent high-throughput sequencing of variable regions of marker genes encoding 16S ribosomal RNA. At the same time, pure cultures of chitinolytic bacteria were isolated.
Three highly active chitinolytic enrichment cultures of microorganisms were obtained: from the copepod homogenate (subclass Copepoda), from worm homogenate Terebellides (Class Polychaeta) and from a seawater sample near the BBS. For the same samples, microbial communities were previously profiled using the 16S rRNA gene. As a result of cultivation on chitin, the composition of communities changed noticeably: bacteria of the genera began to dominate in them Aliivibrio, Photobacterium, Pseudoalteromonas, Cobetia, Endozoicomonas.
Three highly active chitinolytic strains were also isolated, identified by analysis of the 16S rRNA gene as two representatives of the genus Pseudoalteromonas sp. (presumably new species) and Vibrio sp.whose closest relative is Vibrio alginolyticus. All three strains actively degraded chitin and were also capable of growth on other marine polysaccharides—algae components (agar, alginate, carrageenan).
Zones of chitin hydrolysis on Petri dishes with a nutrient medium on which the studied bacteria were placed.
In order to identify chitinase genes (genus Vibrio, Aliivibrio, Pseudoalteromonas, Cobetia, Endozoicomonas) an analysis was carried out of the published genomes of marine microorganisms related to those discovered or isolated during the studies. For some members of the first three genera, the ability to degrade chitin was previously known; for representatives of clans Cobetia And Endozoicomonas such data were not available. Chitinase genes were found in the genomes of representatives Endozoicomonaswhich is consistent with the data of the present study. The ability of bacteria of the genus Cobetia to decompose chitin requires additional research.
“This work is special for us, – speaks Irina DanilovaAssociate Professor, Department of Microbiology, Faculty of Biology, Moscow State University. — On the one hand, we obtained very interesting results on new bacteria producing chitinases, and these bacteria live in northern conditions, and this is a very valuable quality for biotechnology, because in this case all biotechnological processes with proteins obtained from these bacteria can be carried out at low temperatures. This is very important from an economic point of view. On the other hand, all the practical part of the work was done by our students, who received an unforgettable experience of conducting practice-oriented research. We hope that this will give them an additional incentive to stay in science and devote their lives to work aimed at improving human life on our planet. In the future, we will definitely involve our students in other similar studies.”.
The results of the study, supported by a grant from the Ministry of Education and Science No. 075-15-2021-1396 as part of the national project “Science and Universities”, were published in the journal Microbiology.
Based on materials from the press service of Moscow State University. M. V. Lomonosov.
Source: www.nkj.ru